Hi Romy,
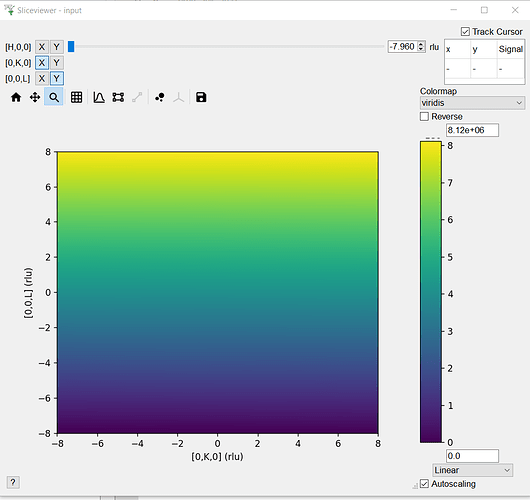
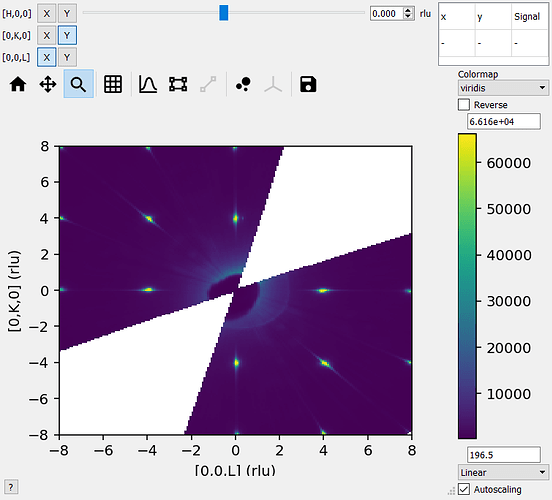
Harriet asked me to look into this - so at least for fake data (see below) the sliceviewer is fine with non-cubic arrays
nh, nk, nl = 3, 5, 10
intens = np.arange(nh*nk*nl)
ws = CreateMDHistoWorkspace(Dimensionality=3,
Extents="-8,8,-8,8,-8,8",
SignalInput=intens,
ErrorInput=np.ones(intens.shape),
NumberOfBins=[nh, nk, nl],
Names=("[H,0,0]","[0,K,0]","[0,0,L]"),
Units="rlu,rlu,rlu",Frames="HKL,HKL,HKL")
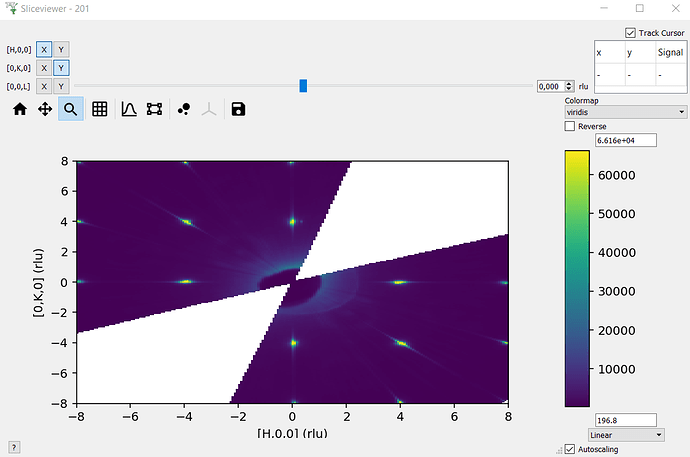
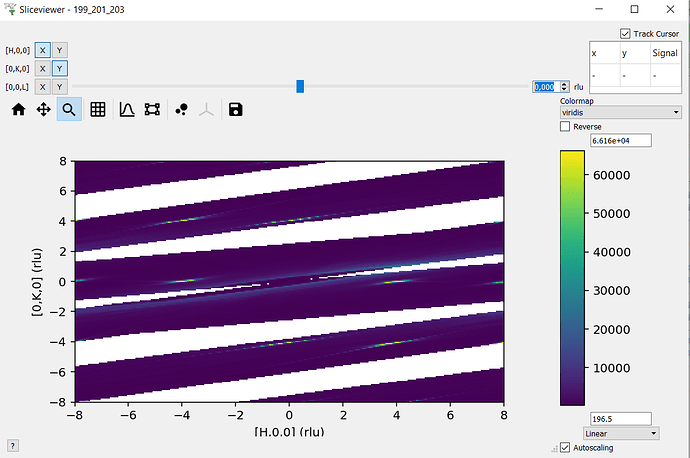
I’m not familiar with how you are rebinning the data (outside of mantid?) - so I think it’s worth checking whether sliceviewer is displaying the data in the workspace correctly or whether it is the rebinning or importing of the data that is causing issues (e.g. reshaping a signal array with wrong row/column order etc.).
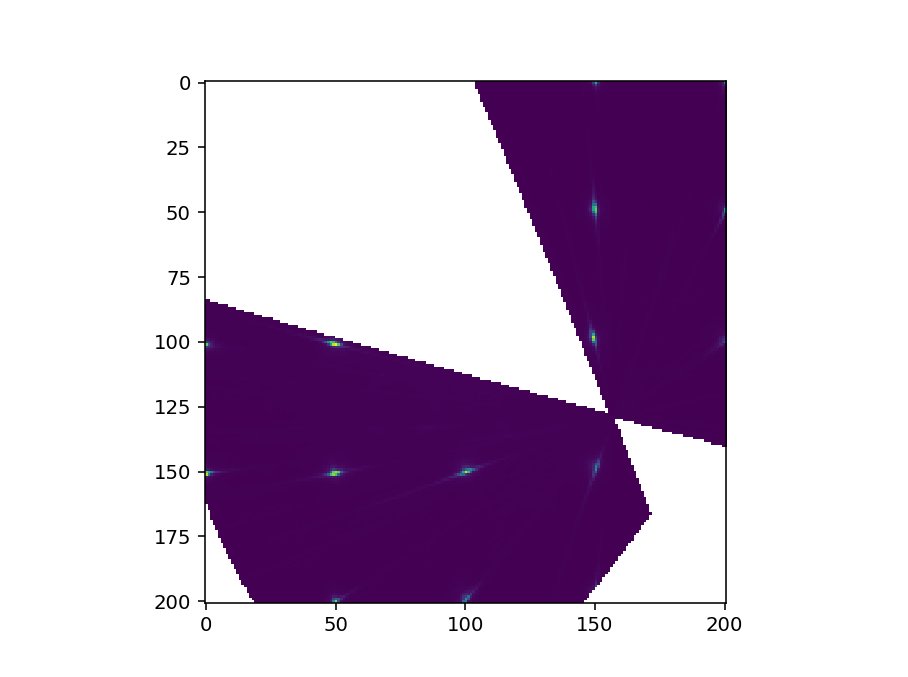
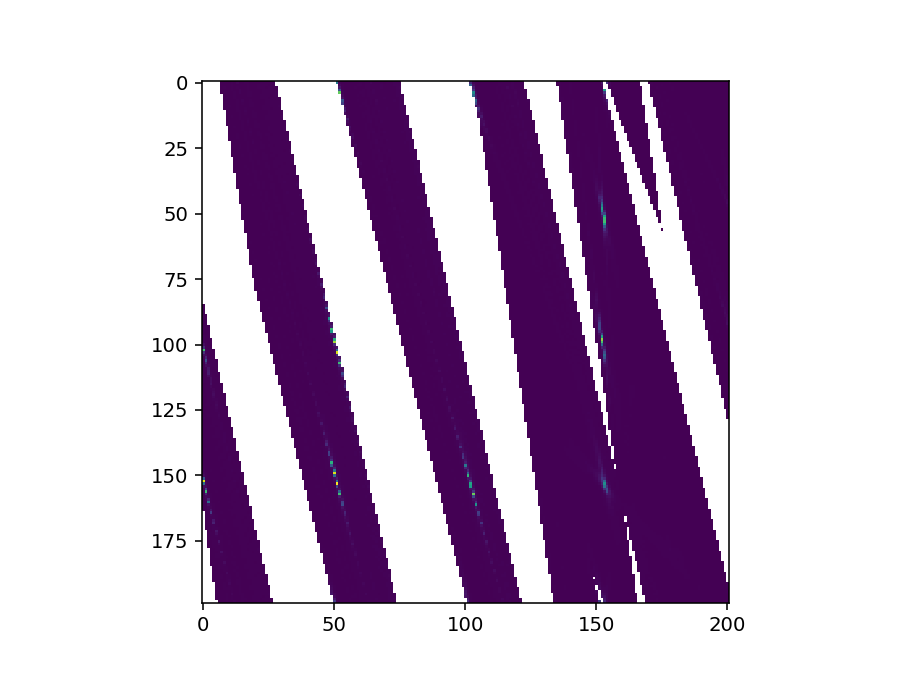
To check this could you just plot the signal array of the workspace like so
fig, ax = plt.subplots()
im = ax.imshow(ws.getSignalArray()[:,:,0])
fig.show()
The image will probably be flipped in some way as I haven’t taken care of the extents (that’s why we have sliceviewer to do the work for us!) - but the image should look recognisably similar to what sliceviewer shows. Could you try this and send a screenshot?
Thanks,
Richard